|
Dinucleotides
- Theoretical Structures
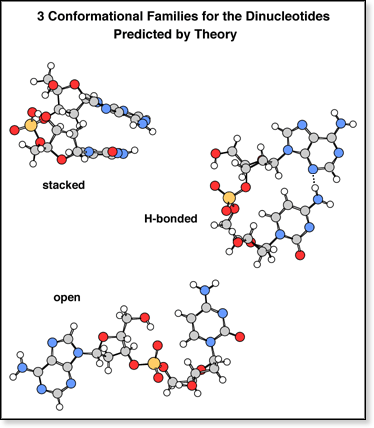 Theoretical
modeling of the dinucleotides (using the simulated
annealing method described in the molecular
mechanics section) predicts three conformational families with similar
energies but significantly different collision cross sections. Examples
of each family (for dAC) are shown at right. Theoretical
modeling of the dinucleotides (using the simulated
annealing method described in the molecular
mechanics section) predicts three conformational families with similar
energies but significantly different collision cross sections. Examples
of each family (for dAC) are shown at right.
The stacked conformers, with the two nucleobases
stacked, are the lowest-energy structures and have the smallest cross
sections. The bases tend to stack so that a carbonyl oxygen on one base
is directly over the NH2 group (or NH on thymine) on the other
base.
The H-bonded conformers, with the two bases
nearly planar with respect to one another and hydrogen bonded to each
other, are the next-lowest-energy structures (by 1-2 kcal/mol) and have
cross sections 10-12 Å2 larger than the stacked
conformers. The H-bonds typically involve the carbonyl oxygen or ring
nitrogen on the 5' base and the amino group on the 3' base. As a result,
no H-bonded conformers are observed for dAT, dCT, or dTT because
the 3' T does not have an amino group and the CH3 group is
usually in the way. For dGT, however, the H-bonds between G and T involve
the NH2 group on the 5' G and the C=O group on the 3' T.
The
open conformers, with the two bases separated from each other,
are the highest-energy structures (by 1-10 kcal/mol) and have cross sections
20-25 Å2 larger than the stacked conformers.
|

