|
Introduction
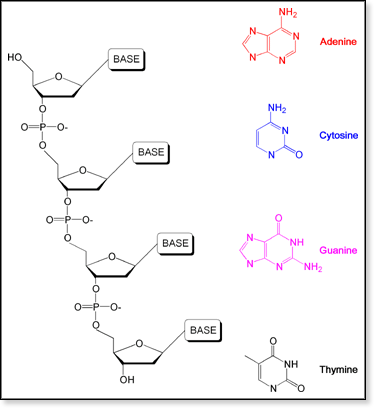 DNA
is a relatively simple polymer composed of repeating nucleotide units.
Each nucleotide contains a phosphate group, a sugar, and a pyridine or
purine base. A simple schematic of a DNA strand and the 4 common bases
are shown at right. DNA
is a relatively simple polymer composed of repeating nucleotide units.
Each nucleotide contains a phosphate group, a sugar, and a pyridine or
purine base. A simple schematic of a DNA strand and the 4 common bases
are shown at right.
Hydrogen
bonding between complimentary bases on two DNA strands (A·T and
C·G) leads to duplex formation and the famous double-helix structure
described by Watson and Crick. Additional studies, however, have shown
that DNA actually forms a variety of helical structures and can even form
triplexes and quadruplexes. These various helical structures are the result
of variations in the structures of the nucleotides (in particular, the
conformation of the sugar and the relative positions of the bases with
respect to the sugars) [Sinden, R.R. DNA Structure and Function;
Academic Press: San Diego, 1994]. The transition from a right-handed "B"
helix is accompanied by a change in the sugar pucker and a rotation of
a base. A change in sugar pucker also accompanies the transition of a
"B" helix into a more compact "A" helix. Protonation
of adenine (A) leads to the formation of A+·C and A+·G
base pairs, instead of the "normal" A·T pairs. Protonation
of cytosine (C) is essential for the stabilization of triplexes.
Most
structural studies of DNA are performed in the condensed phase using X-ray
crystallography or multidimensional NMR analysis. However, recent advances
in ionization sources (such as MALDI and ESI) have led to an increase
in the number of gas-phase studies on DNA using mass spectrometry. There
has also been an increase in the number of studies involving the theoretical
modeling of DNA structures. Gas-phase experiments are ideal for direct
comparison with these theoretical investigations.
We
have used mass spectrometry and ion mobility methods to investigate the
conformational and energetic properties of a series of mono-, di-, and
trinucleotides. Questions concerning structural differences between protonated
and deprotonated nucleotides, base-base interactions, and zwitterion formation
are addressed.
- Mononucleotide
Structures
- Hydration
of Mononucleotides
- Dinucleotides
- Trinucleotides
- Duplex
Structures
- Quadruplex
Structures
Bowers group members who have worked on these projects include
|


